library("palmerpenguins")
library("rpart")
library("rpart.plot")
library("tidymodels")Years ago, I would use the caret package to perform a random forest search and plot an example of a decision tree. Can we do that now in the TidyModels module?
Here I am adapting code from Stack Overflow
df <- penguins |>
mutate(species = factor(species))
data_split <- initial_split(df)
df_train <- training(data_split)
df_test <- testing(data_split)df_recipe <- recipe(species ~ ., data = df) %>%
step_normalize(all_numeric())#building model
tree <- decision_tree() %>%
set_engine("rpart") %>%
set_mode("classification")#workflow
tree_wf <- workflow() %>%
add_recipe(df_recipe) %>%
add_model(tree) %>%
fit(df_train) #results are found here tree_fit <- tree_wf |>
extract_fit_parsnip()
rpart.plot(tree_fit$fit, roundint = FALSE)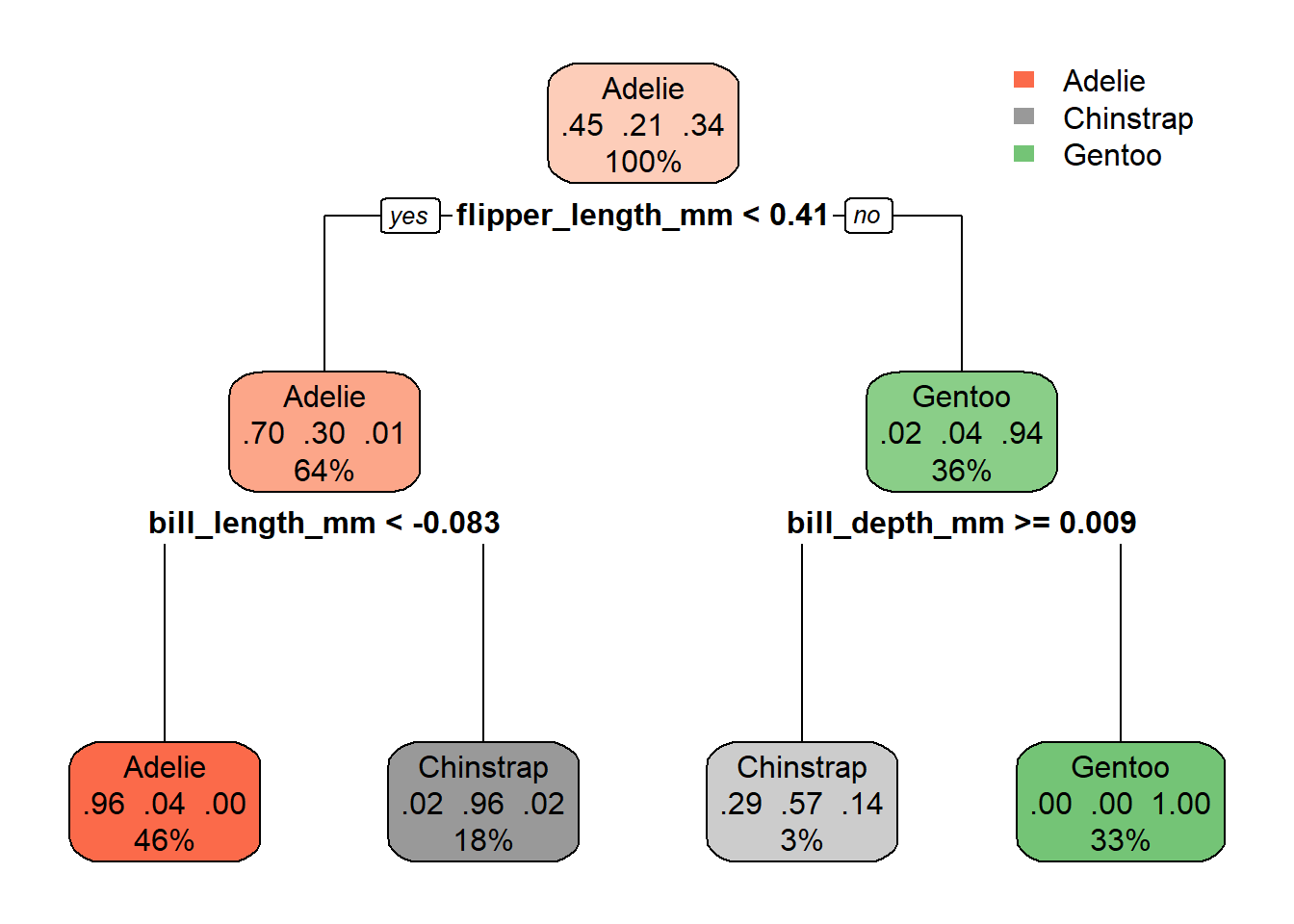
So far,
- response variable has to be a factor type—and hence should be categorical—in a classification setting
- I need to learn what all of those numbers mean!
sessionInfo()R version 4.2.2 (2022-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19044)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] yardstick_1.2.0 workflowsets_1.0.1 workflows_1.1.3
[4] tune_1.1.1 tidyr_1.3.0 tibble_3.2.1
[7] rsample_1.1.1 recipes_1.0.6 purrr_1.0.1
[10] parsnip_1.1.0 modeldata_1.1.0 infer_1.0.4
[13] ggplot2_3.4.2 dplyr_1.1.2 dials_1.2.0
[16] scales_1.2.1 broom_1.0.4 tidymodels_1.1.0
[19] rpart.plot_3.1.1 rpart_4.1.19 palmerpenguins_0.1.1
loaded via a namespace (and not attached):
[1] jsonlite_1.8.4 splines_4.2.2 foreach_1.5.2
[4] prodlim_2023.03.31 GPfit_1.0-8 yaml_2.3.7
[7] globals_0.16.2 ipred_0.9-14 pillar_1.9.0
[10] backports_1.4.1 lattice_0.20-45 glue_1.6.2
[13] digest_0.6.31 hardhat_1.3.0 colorspace_2.1-0
[16] htmltools_0.5.4 Matrix_1.5-3 timeDate_4022.108
[19] pkgconfig_2.0.3 lhs_1.1.6 DiceDesign_1.9
[22] listenv_0.9.0 gower_1.0.1 lava_1.7.2.1
[25] timechange_0.2.0 generics_0.1.3 ellipsis_0.3.2
[28] withr_2.5.0 furrr_0.3.1 nnet_7.3-18
[31] cli_3.6.1 survival_3.4-0 magrittr_2.0.3
[34] evaluate_0.21 future_1.32.0 fansi_1.0.4
[37] parallelly_1.35.0 MASS_7.3-58.1 class_7.3-20
[40] tools_4.2.2 data.table_1.14.8 lifecycle_1.0.3
[43] munsell_0.5.0 compiler_4.2.2 rlang_1.1.0
[46] grid_4.2.2 iterators_1.0.14 rstudioapi_0.14
[49] htmlwidgets_1.6.2 rmarkdown_2.21 gtable_0.3.3
[52] codetools_0.2-18 R6_2.5.1 lubridate_1.9.2
[55] knitr_1.42 fastmap_1.1.1 future.apply_1.10.0
[58] utf8_1.2.3 parallel_4.2.2 Rcpp_1.0.10
[61] vctrs_0.6.1 tidyselect_1.2.0 xfun_0.39